-
General Information -
How to Use: Example with Metaraminol -
Glossary -
References -
Problem In Running Application?
Example With Metaraminol
Step 1) Draw the structure of Metaraminol
First we need to draw the structure of Metaraminol. JSME molecule sketcher JSME is provided in browser.
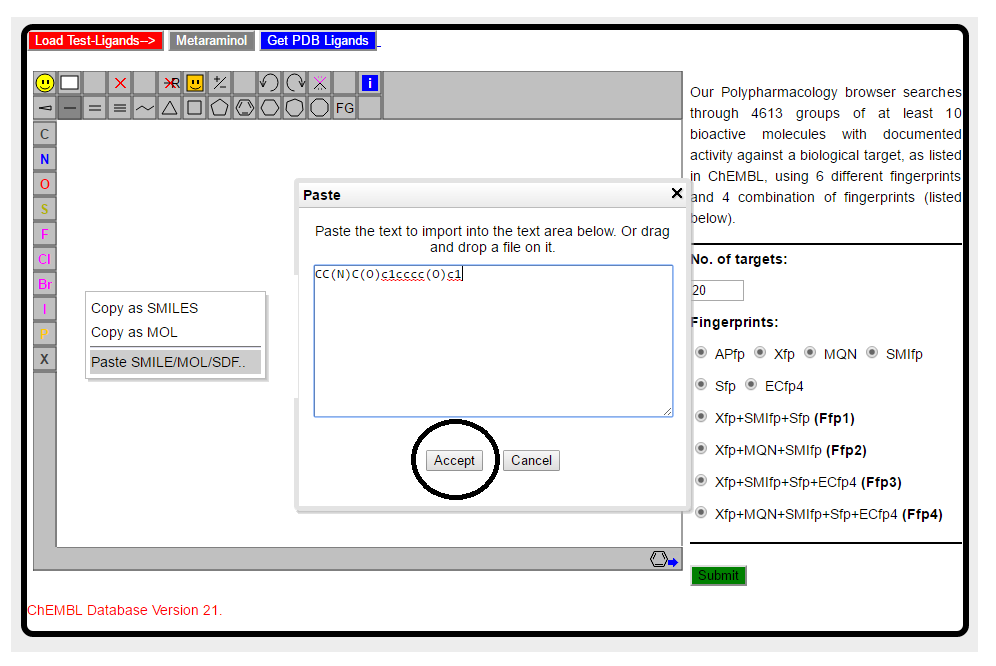
In case you have smiles notation of molecule, option is available to paste the smiles in JSME sketch window. Right click in the sketch window and paste the smiles of Metaraminol CC(N)C(O)c1cccc(O)c1 as shown below
.
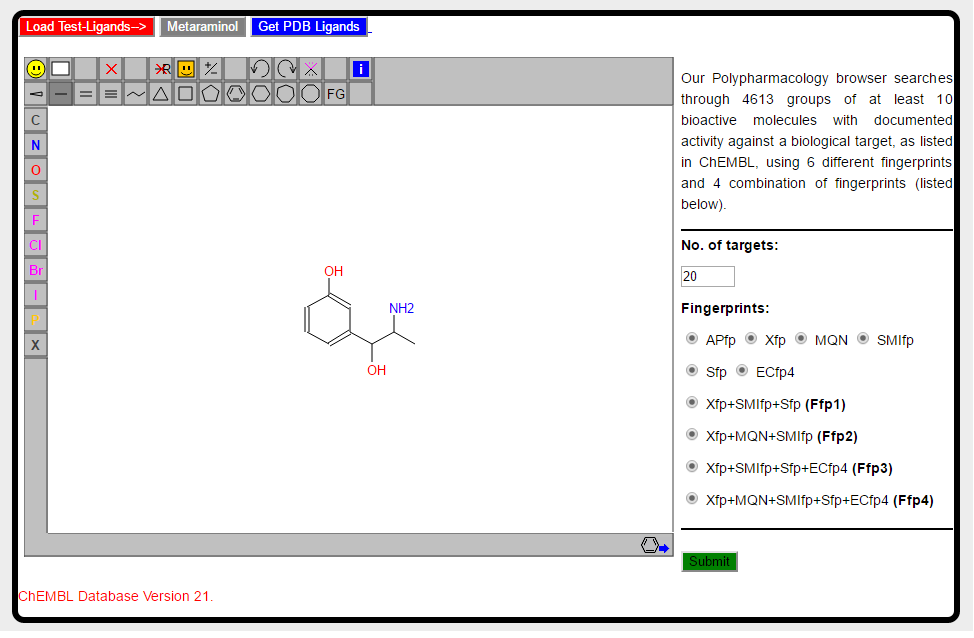
Alternatively you can use the Metaraminol button located on the upper part of sketch window, which will automatically load the molecule for you. Shown below is JSME sketch window with the structure of Metaraminol.
Step 2) Submit the job and wait for Result
Once the structre of query molecule is drawn, we are ready to submit the job. Hit the green "Submit" button.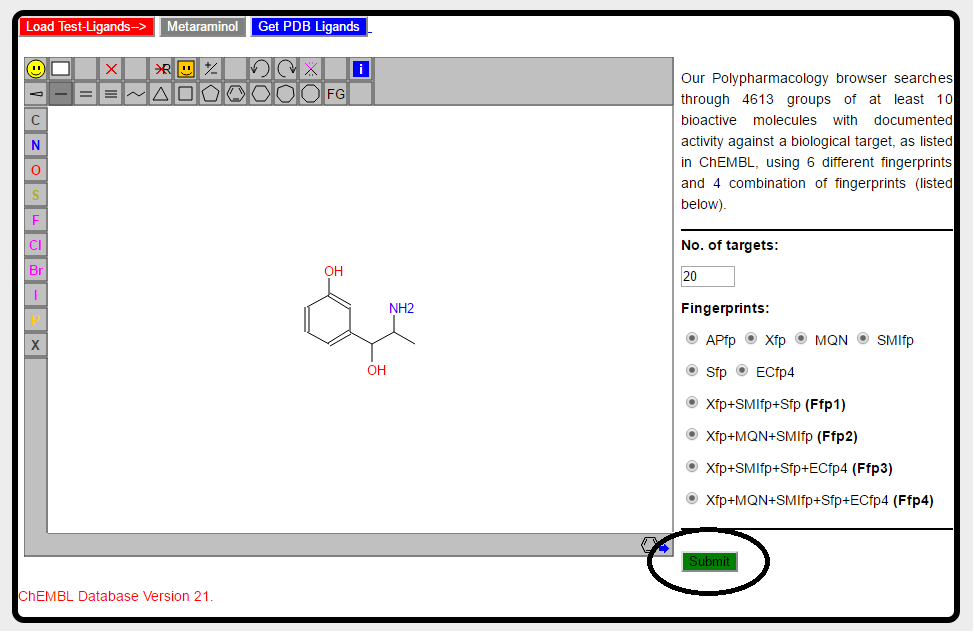
PPB: Polypharmacology Browser searches using 6 different fingerprints (Xfp, MQN, SMIfp, APfp, Sfp and ECFP4) and 4 combinations of fingerprints (Hyp1-Hyp4). Search procedure usually takes maximum of 1 minute. After submitting the job new window will appear which will ask you to wait.
Step 4) Result Window
Result window is shown below.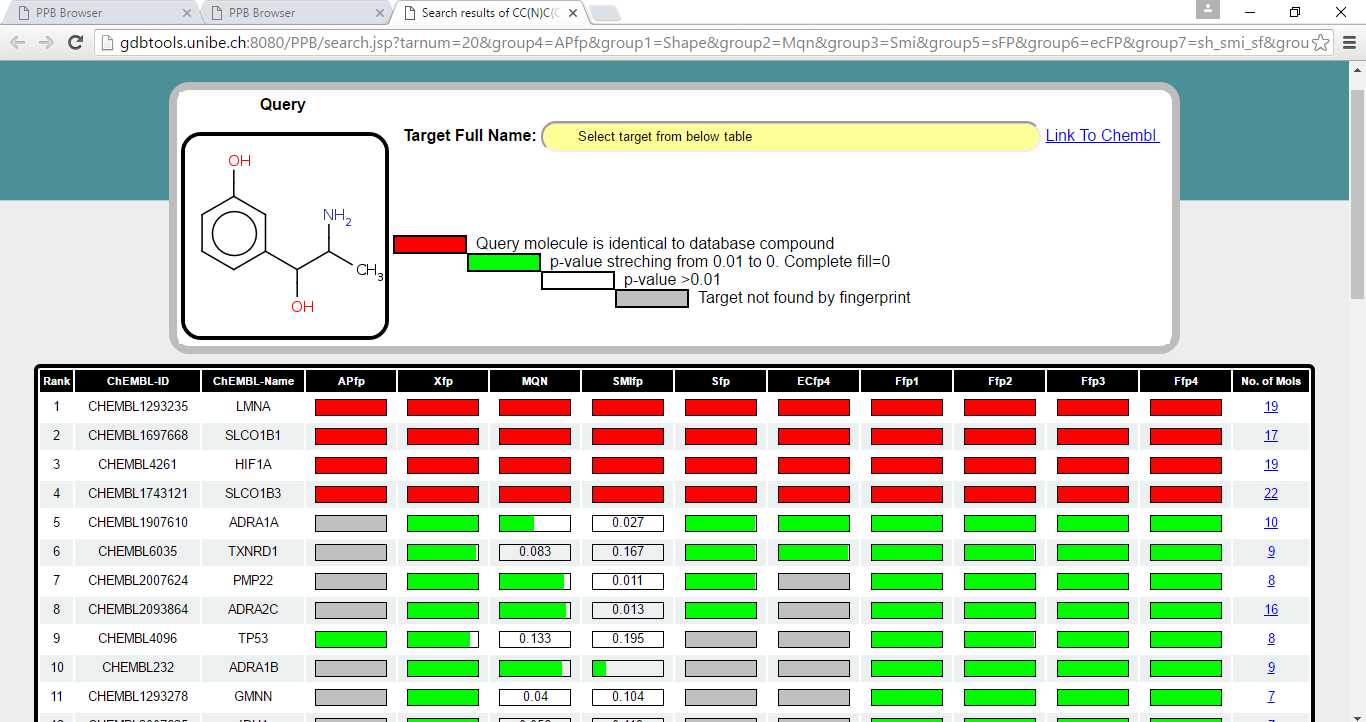
Step 4 a) Target Table
This table list the probable target proteins for query Metaraminol. The targets are retrieved using 6 different fingerptints and 4 combination of fingerprints independently. Each of the fingerprint sort the database according to decreasing similarity to Metaraminol and collect the compounds from sorted database till it find 20 targets.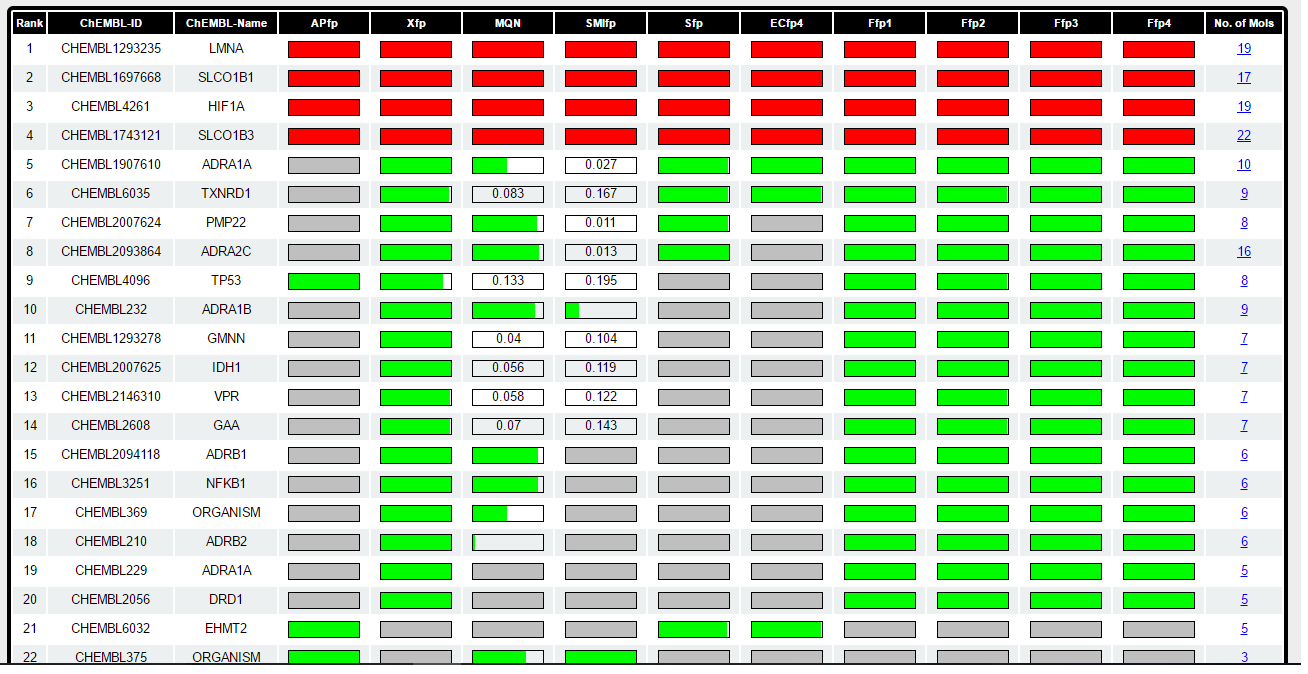
Target are sorted according to the their appearence in different fingerprint spaces followed by sum of p-value across all fingerprint spaces. It means if the target is found by all the fingerprint it will appear top in the list. Bar in front of each target represent the probability which stretch from 0.01 to 0. When the PValue is 0 bar is completely fill with green color and when PValue is more than 0.01 its completly empty and indicated by the corrosponding PValue. When the target is not found by fingerprint the bar is filled with grey color. Calculation of PValue is explained in detail in "General Information" section (also see below). For each of the target number of already known bio-active molecules found by browser is shown in last column in table. This number is total across all the fingerprints. When the same compound is retreived by all the fingerprints for perticular target protein and when it is identical to input query molecule probability bar is marked in red color (for e.g in 1-3 targets in table above).
Step 4 b) Looking for target full name
Clicking on the row in target table will shows you the full name of target in the panel located above the target table. For instance full name for ADRA1A (row 5 selected) is shown below.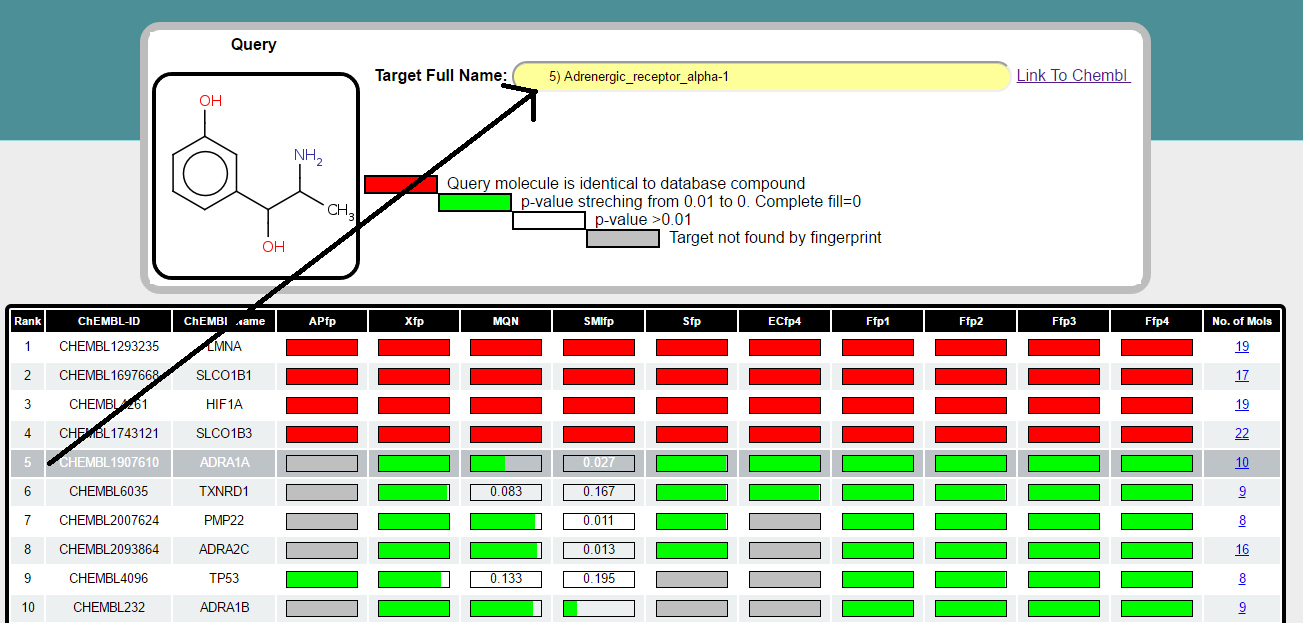
Step 4 c) ADRA1A: Connect to ChEMBL
Once table row corrosponding to ADRA1A is selected, one can click on Link To ChEMBL hyperlink to get details information about target from ChEMBL.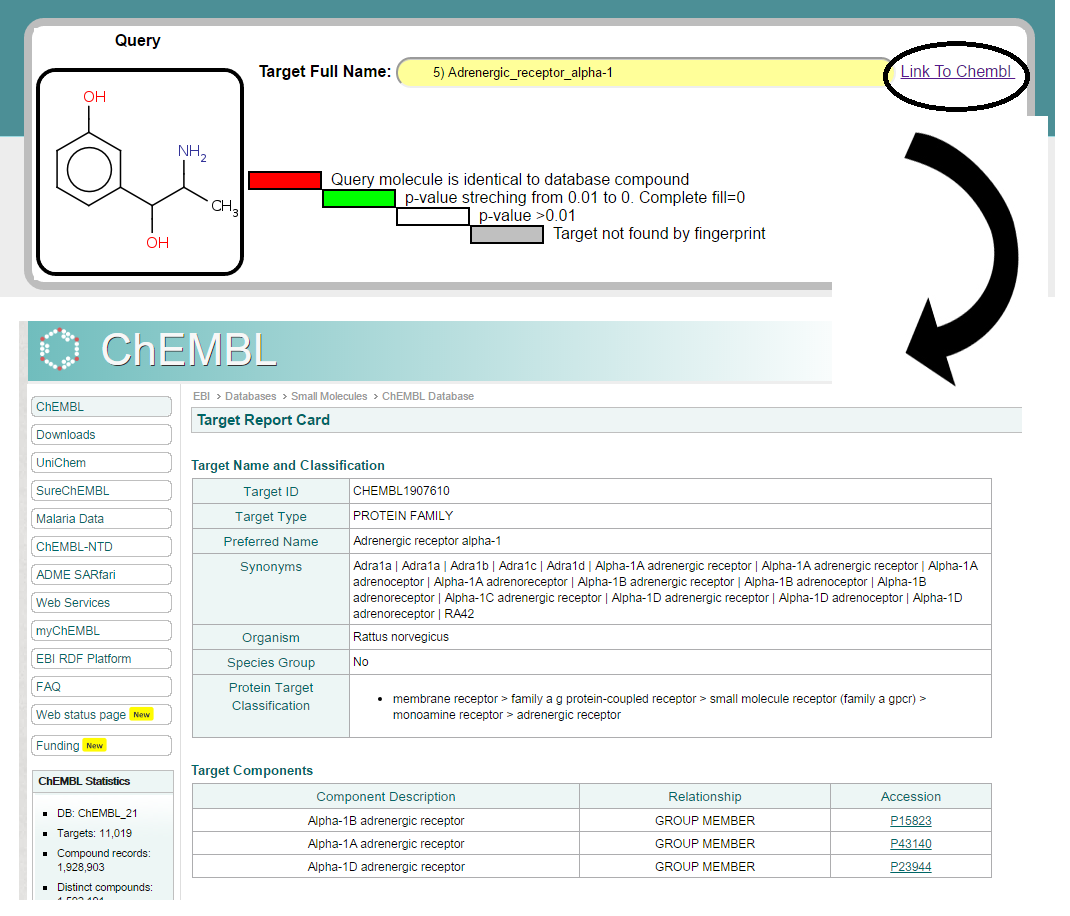
Step 4 d) ADRA1A: View Molecules
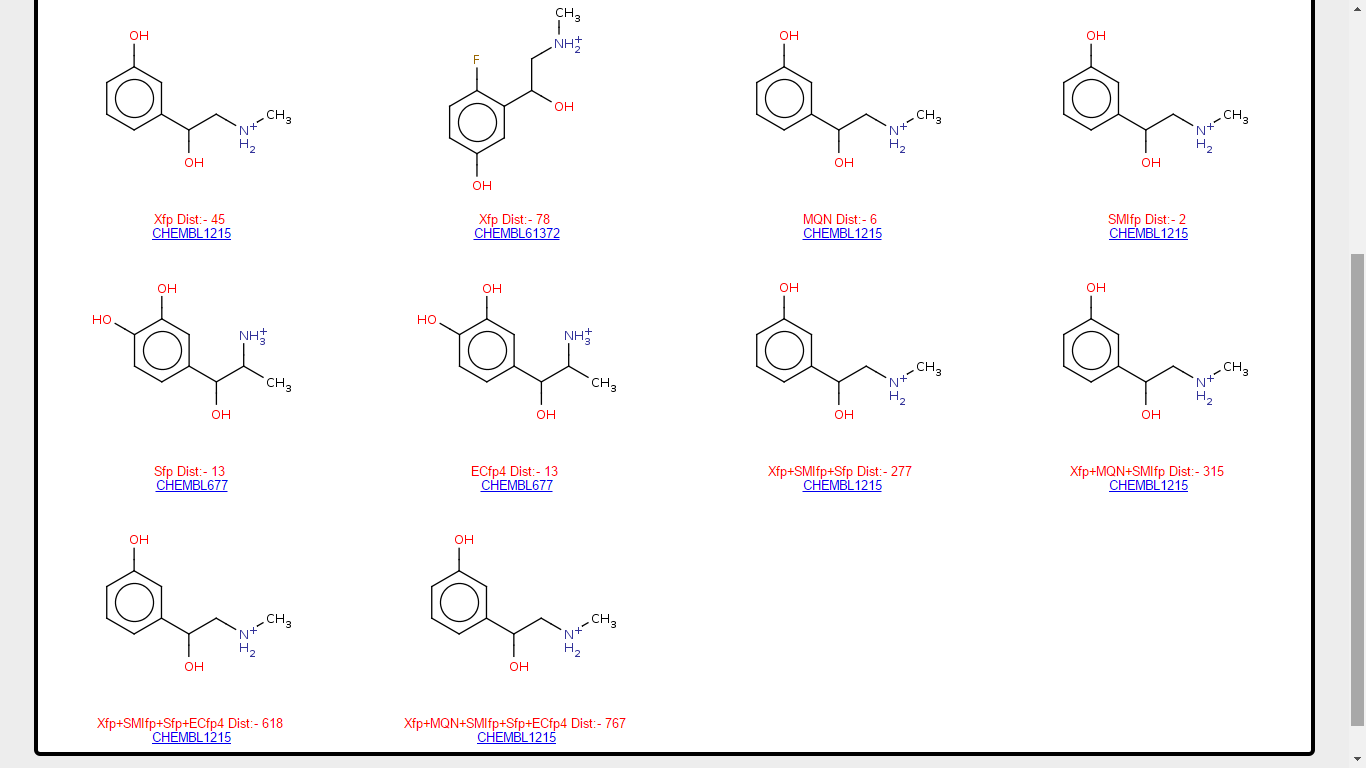
One can look at the molecules collected by browser to predict the ADRA1A as target protein of Metaraminol. In the Target table click on the No. of Molecules at row 5, which will redirect you to the list of molecules retrieved by browser and are already reported for ADRA1A target. Molecules are tag with the name of fingerprint (which found this molecules), city block distance for the molecule with respect to Metaraminol and ChEMBL ID of compound which will direct the user to ChEMBL database to have a additional information for this compound.